IT-4-P-5814 Development of "Adaptive SEM" Technology for in situ Genome/Proteome Expression Analysis in Single Cell Level
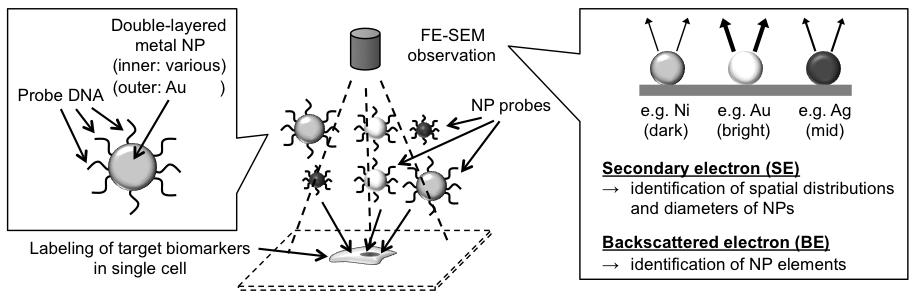
Obtaining expression information in a cellular system is essential for understanding the mechanisms of living systems. One useful way is in situ measurement of expressed biomarkers in single cell level using a lot labels; however, production and identification of such labels are still challenging. We propose a new sensing technology based on the field emission scanning electron microscopy (FE-SEM), which is a comprehensive development of production and identification of nano-particle (NP) labels for simultaneous in situ measurements of expressed biomarkers in single cell (Fig.1).
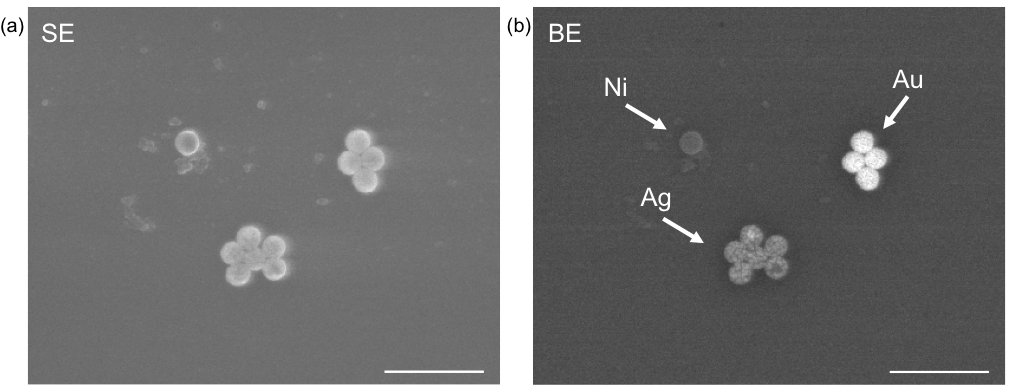
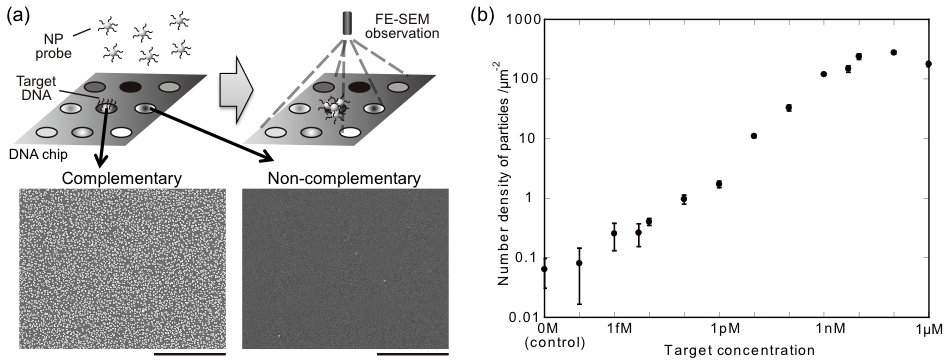
For the fabrication of NPs, various sizes of polystyrene spheres were used as templates, and metals were deposited on the spheres by thermal evaporation. By using this method, more than 500 types of NPs were fabricated. Metal shell layers were formed by thermal evaporation; therefore, multi-layered NPs can be fabricated with sequential evaporation. We used double-layered NPs; outer is Au for easy immobilization of biomolecules to use these NPs as labeling probes, and inner layer is various to apply label varieties in FE-SEM observation. In this study, probe DNAs were immobilized onto the outer Au layers (referred to as "NP probe" hereafter), and target DNAs on a substrate were reacted with the NP probes as a model. For its detection, FE-SEM observations were performed to identify numbers and elements of hybridized NP probes on the substrate. Spatial distributions and diameters of NP probes were identified by secondary electron (SE) observation, and elements of NP probes were identified by backscattered electron (BE) observation as the difference of intensities in the BE image caused by the difference of atomic number of inner metal layer (Fig.2). In results, six different elements were simultaneously distinguished by BE observation [1], indicated that targets can be simultaneously labeled and identified with high spatial resolution by the combination of NP probe labeling with BE image analysis. In addition, detection sensitivity of target DNA in this method was femto-molar order [2] (Fig.3), which is 1,000 times higher than that in conventional fluorescent labeling and optical detection, indicated that our method is suitable for the detection of a few biomarkers in single cell. We call it "adaptive SEM" technology (i.e., NP identification is "adaptive" for various targets). These results indicate a possibility for quantitative in situ detection of expressed biomarkers in single cell level by the suggested technology based on NP probe labeling and FE-SEM identification.
References
[1] Kim, H., Negishi, T., Kudo, M., Takei, H., Yasuda, K., J. Electron Microsc. 59 (5), 379-385 (2009)
[2] Kim, H., Kira, A., Yasuda, K., Jpn. J. Appl. Phys. 49 (6), 06GK07, 1-7 (2010)
We thank Ms. M. Murakami and Ms. M. Naganuma for their technical assistance. This work was financially supported by the Japan Prize Foundation, JSPS, and KAST.