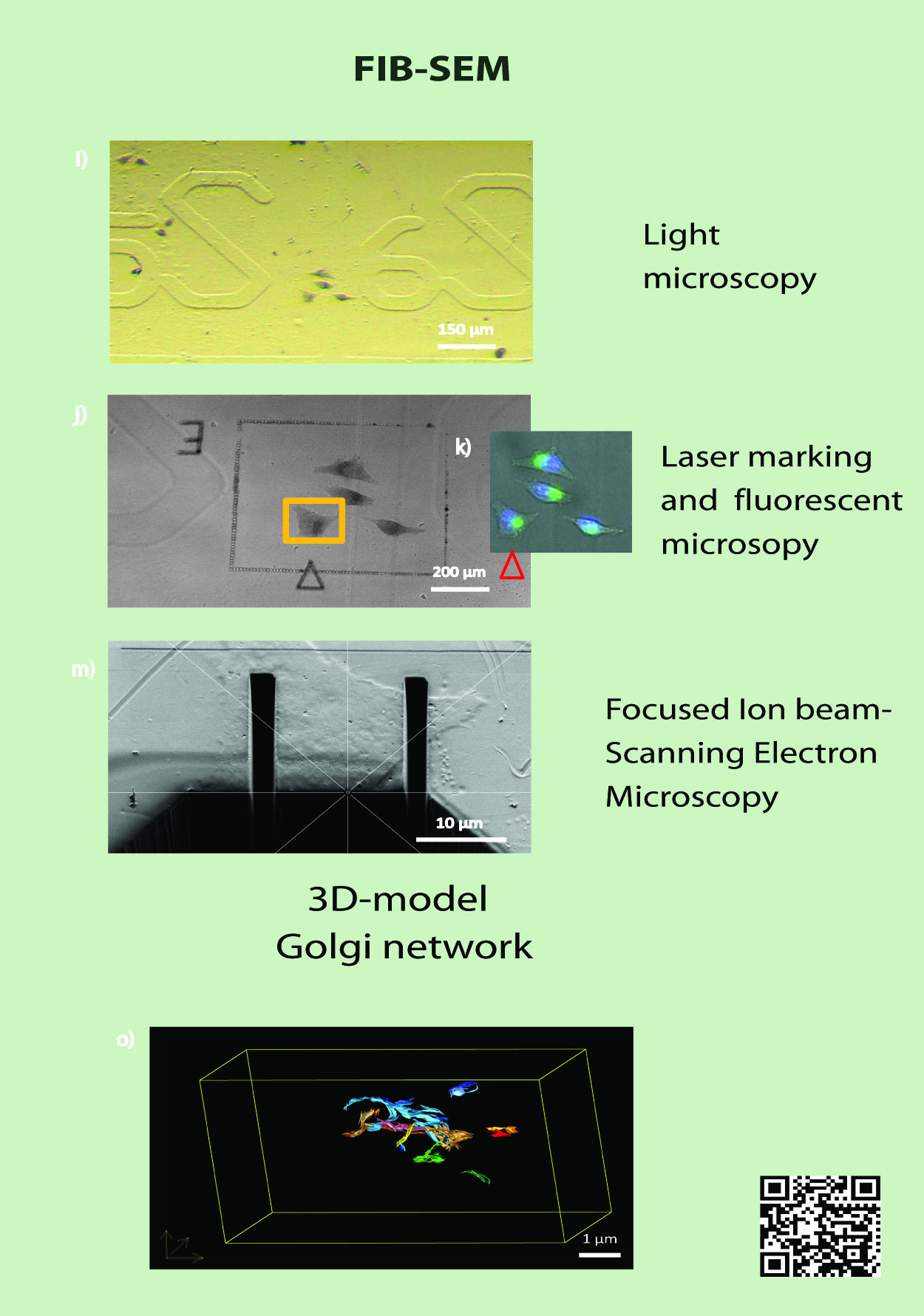
IT-13-P-2874 Targeted 3D-CLEM workflow on cultured cells
Correlative light and electron microscopy (CLEM) experiments uniquely provide a highly accurate link between the imaging of living cells and their 3D ultrastructure (4). However, CLEM generally suffers from a low throughput. The major hurdles include tracking the object throughout the different imaging modalities, the tedious procedures for sample preparation and the lack of automation in the data acquisition by electron microscopy. We aim to overcome these issues by developing an automated correlative workflow that links live cell imaging to high resolution 3D electron microscopy using FIB-SEM. The approach we will develop enables collecting statistically significant data from a large number of cells in a heterogeneous population, clearing the way to statistical analysis of important mechanisms in cell biology. Aiming to develop a flexible and versatile approach, we foresee applications of our method in other biological areas such as pharmacology, developmental biology or virology.
Currently, our workflow is developed and tested on cultured cells and employed to in a high-throughput study of Golgi apparatus organization. In a tight collaboration with the team of R. Pepperkok (EMBL Heidelberg), we visualize at the ultrastructural level how specific mutations influence the morphology of the Golgi apparatus. Using an automated light microscopy platform, large genome wide siRNA knockdown screens have led to the identification of key genes for the morphogenesis and function of the Golgi apparatus (3). Light microscopy was utilized to screen for specific phenotypes, fluorescent microscopy to find cells of interest and electron microscopy to look at the ultrastructure.
References:
1) Briggman K. L. and D. D. Bock (2012). "Volume electron microscopy for neuronal circuit reconstruction" Curr Opin Neurobiol 22(1): 154-161.
2) Colombelli J., et al. (2008). “A correlative light and electron microscopy method based on laser micropatterning and etching.” Methods Mol Biol. 457:203-213
3) Simpson J. C., et al. (2012). "Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway" Nat Cell Biol 14(7): 764-774.
4) Spiegelhalter C., et al. (2010). “From dynamic cell imaging to 3D ultrastructure: novel integrated methods for high pressure freezing and correlative light-electron microscopy” PLOS One 5(2): 203-213
5) Villinger C., et al. (2012). "FIB/SEM tomography with TEM-like resolution for 3D imaging of high-pressure frozen cells" Histochem Cell Biol 138(4): 549-556.
Many thanks to Team Schwab, Team Pepperkok and our collaboration partner Zeiss.