IT-10-O-2543 On-axis electron tomography of needle-shaped biological samples
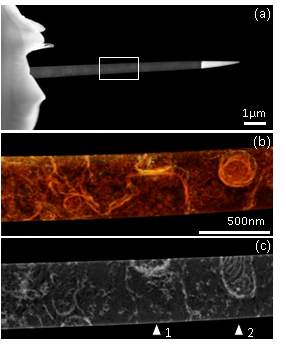
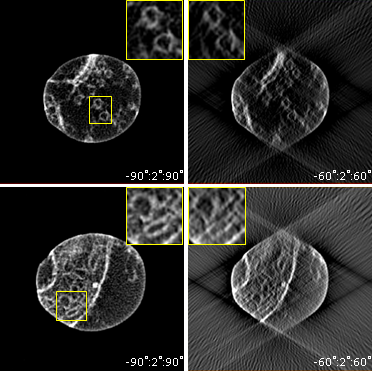
A key challenge in structural biology is to image large volumes while maintaining sufficient resolution to identify small features in their original cellular context [1]. Electron tomography (ET) has contributed greatly to this field, but imaging sections thicker than a few hundred nanometers is difficult because of the sample geometry and microscope configuration: as the specimen is tilted to high angles, the thickness increases and the quality of the images deteriorates. Moreover, for geometric constraints, the tilt range rarely exceeds ±70°, leading to elongation and blurring of features, and an overall challenging volume to segment. Here, we show that preparing a needle-shaped sample rather than a flat section can alleviate many of the limitations encountered in biological ET. The technique is illustrated on a 500nm diameter needle extracted from an epoxy-embedded portion of the nucleus accumbens shell from a rat brain. The sample was prepared in a Helios NanoLab focused ion beam (FIB) machine and transferred to an on-axis tomography holder. An HAADF-STEM tilt series was then acquired from -90° to +90° with 1° increment, using an FEI TITAN microscope operating at 300kV, and Inspect3D was used for the alignment and reconstruction by weighted backprojection. Figure 1(a) shows a low magnification view of the needle and the region selected for tomography. A voxel projection and slice through the reconstructed needle (b,c) provide highly detailed structural information. In Figure 2, we compare the quality of ET results from -90°:2°:+90° and -60°:2°:60° acquisitions. Cross-sections through a portion of exitatory synapse and mitochondrion (positions 1 and 2 in Figure 1(c)) illustrate the advantages of a full tilt range on-axis ET experiment with enhanced signal-to-background ratio and isotropic sharpness of features. Note that the ±60° cylindrical volume shown here is still of better quality than a reconstructed section from similar tilt range, since the thickness remains constant throughout the tilt series.
Combining this novel sample preparation technique with advanced imaging modes (BF-STEM for example [2]) and sophisticated reconstruction algorithms such as compressed sensing [3], we anticipate that ET will provide a complementary method to serial sectioning and FIB-SEM slice-and-view techniques [4].
[1] W. Baumeister, Current Opinion in Structural Biology 2002, 12(5):679.
[2] A.A. Sousa et al., Journal of Structural Biology 2011, 174(1): 107.
[3] Z. Saghi et al., Nano Letters 2011, 11(11):4666.
[4] Samples provided by Andrea Falqui and Roberto Marotta, IIT, Genova, Italy.
The research leading to these results has received funding from the European Union Seventh Framework Programme under Grant Agreement 312483-ESTEEM2 (Integrated Infrastructure Initiative–I3), as well as from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC grant agreement 291522-3DIMAGE.