IT-13-P-2498 FIB/SEM tomography for 3D visualization of virus infected fibroblasts with TEM-like resolution.
Keywords: FIB/SEM tomography, high pressure freezing, freeze substitution
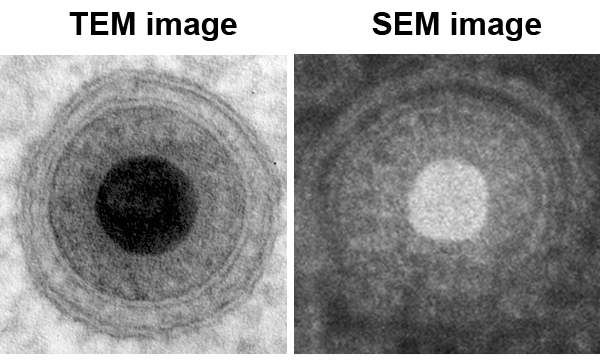
Focussed ion beam/scanning electron microscopy (FIB/SEM) tomography is a novel electron microscopy technique that is increasingly used within life sciences. Here we present its application for ultrastructural visualization of fibroblasts infected with human cytomegalovirus (HCMV). For that we employed optimized sample preparation protocols including high pressure freezing and freeze substitution. The result was an improved ultrastructural preservation and a high image contrast. Additionally, our well established embedding procedure of cell monolayers in Epon allows not only ultrathin sectioning but also sample preparation for FIB/SEM tomography. The detection of the secondary electron signal allows us to resolve cellular and viral ultrastructures down to the level of lipid bilayers (Fig. 1).
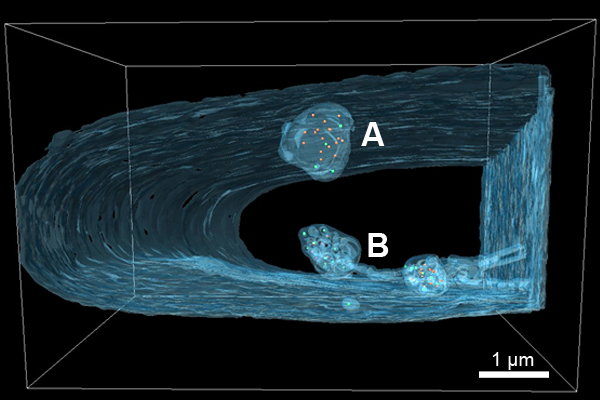
The main focus of our work is the egress of HCMV capsids from the nucleus (capsid formation and genome packaging) into the cytoplasm (further virion maturation). The diameter of virus capsids exceeds the diameter of nuclear pores. Hence, the virus has to find an alternative path to leave the nucleus. It is known that this process is made possible by an envelopment and de-envelopment process at the inner (INM) and outer nuclear membrane (ONM), respectively [1, 2]. We visualized a portion (z=5 µm) of an HCMV infected nucleus with FIB/SEM tomography. The resulting 3D reconstruction showed that the surface area of the INM was increased through large infoldings which can extend deep into the nucleoplasm (Fig. 2). DNA free and DNA filled capsids were both present within these infoldings (perinuclear capsids). This model has already been postulated based on 2D TEM images [3]. Our 3D data now confirm this model. Additionally, the slice thickness of 20 nm allowed imaging of every nuclear as well as perinuclear capsid within the analyzed volume. This gives the opportunity to analyze the capsid distribution in 3D, thus, making the results and interpretation of 2D TEM images more accurate. In conclusion, our FIB/SEM data provide a detailed image of the nuclear stages of HCMV morphogenesis, from capsid assembly and DNA packaging to capsid egress.
[1] Skepper JN et al. (2001).J. Virol. 75, 5697–5702.
[2] Mettenleiter TC et al. (2013). Cell. Microbiol. 15, 170–178.
[3] Buser C et al. (2007). J. Virol. 81, 3042–3048.