IT-10-P-1866 A memory efficient method for 3D object reconstruction with HAADF STEM depth sectioning
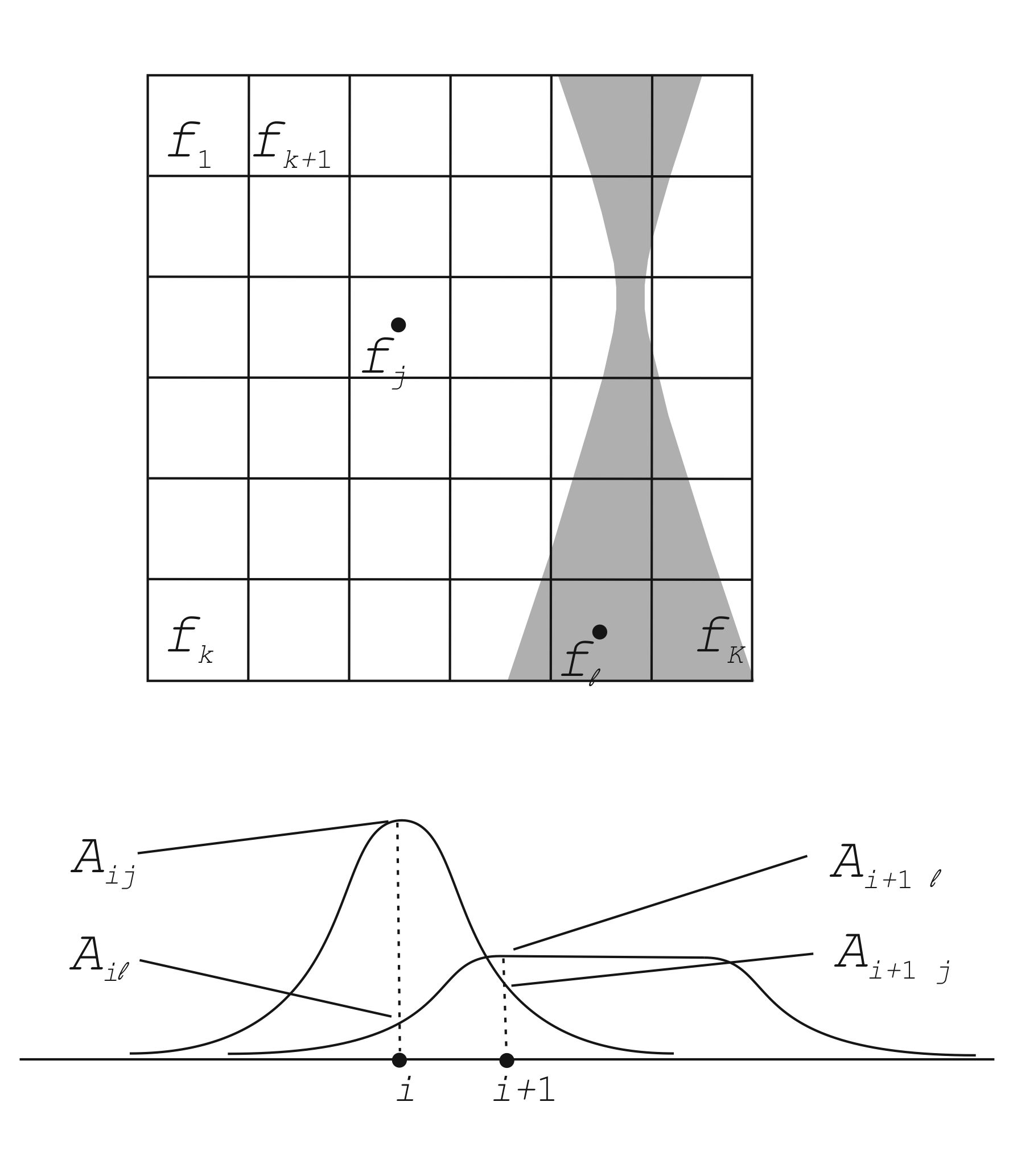
In high angle annular dark field scanning transmission electron microscopy (HAADF STEM) depth sectioning, the object is illuminated with a beam with a small depth of field, caused by a large beam convergence angle, see Fig. 1.
In [Ultram. 110 (2010) 548--554], it is shown that the simultaneous iterative reconstruction technique (SIRT) applies to depth sectioning. The SIRT algorithm is given as
fk+1 = fk + AT [ ( q - A fk ) / A If ] / [ AT Ip ],
where k indicates the iteration number, A is the projection matrix, f is the object vector and q the experimental projection, arithmetic operators between vectors are elementwise and Ip and If each denote a vector of which each element equals 1 with a length equal to that of the projection and the object, resp.
The matrix A scales badly with object size and readily grows too large for the computer memory. Here, we propose to perform the matrix-vector multiplication A fk implicitly through a 2D convolution of each horizontal layer of the object with its corresponding single atom image, followed by a sum in the vertical direction over all horizontal layers. Implementation of the matrix-vector multiplication A If is analogous. The multiplication of AT with the vector ( q - A fk ) / A If can be carried out implicitly by stacking the image in an 3D array and convolving each of the layers with the corresponding single atom image. The matrix-vector multiplication AT Ip can be implemented analogously. The memory load is now reduced to storing the object.
A technique analogous to charge flipping [Acta Cryst. A64 (2008) 123--134] is added: After each iteration values below a certain positive threshold have their sign reversed.
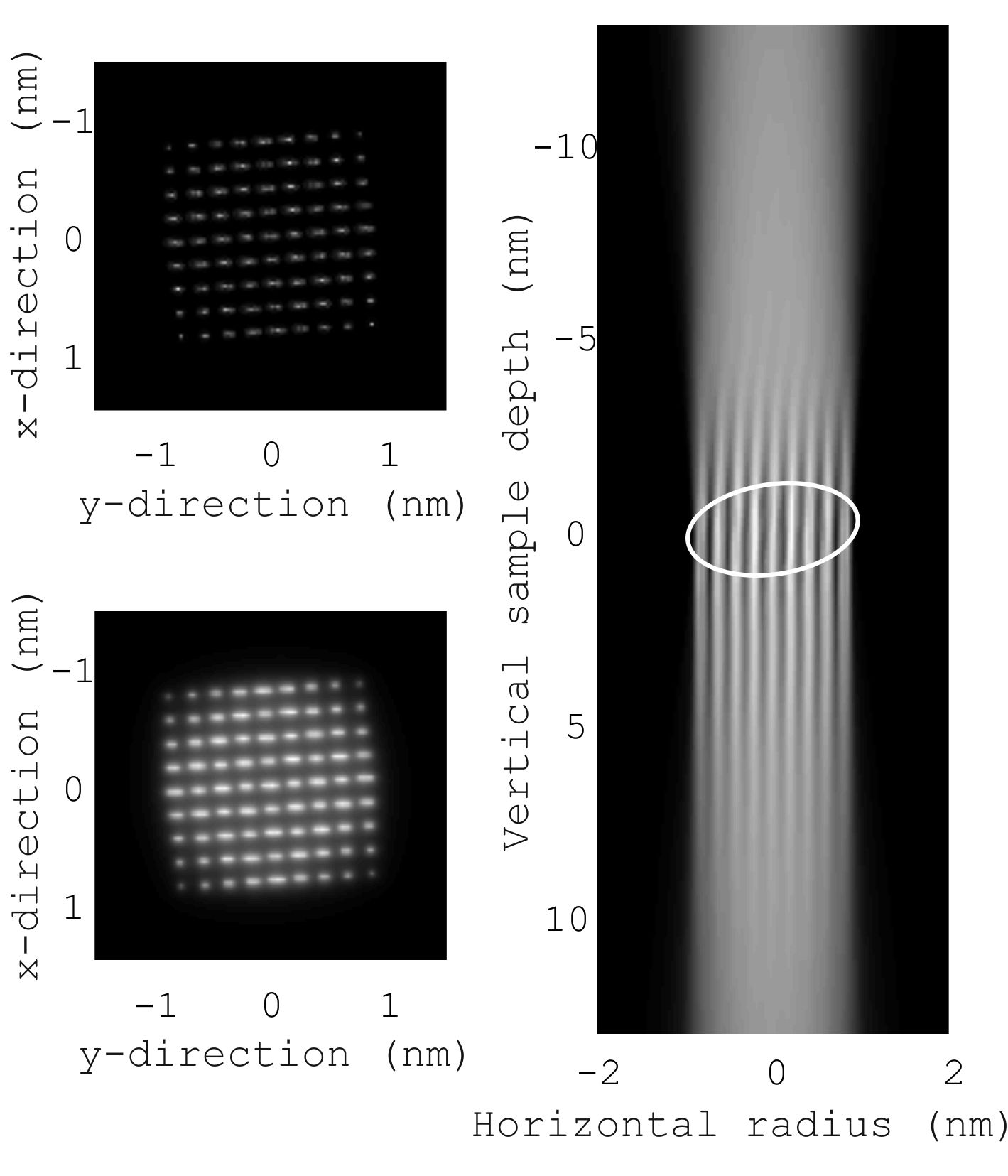
The validity of these implicit matrix-vector multiplications is tested with a simulation of a 1.6nm Au particle. The microscope parameters are: Acceleration voltage: 200kV; C1: -2.67nm; C3: 3.54μm; C5: -1.13mm; C7: 10cm; convergence semi-angle: 86.8mrad. The object measures 1000 x 1000 x 125 voxels of 8 x 8 x 210 pm3. See Fig. 2. The particle is tilted away from the zone-axis. The single atom images encoded in the matrix A are a convolution of the Au atom potential and the probe intensity.
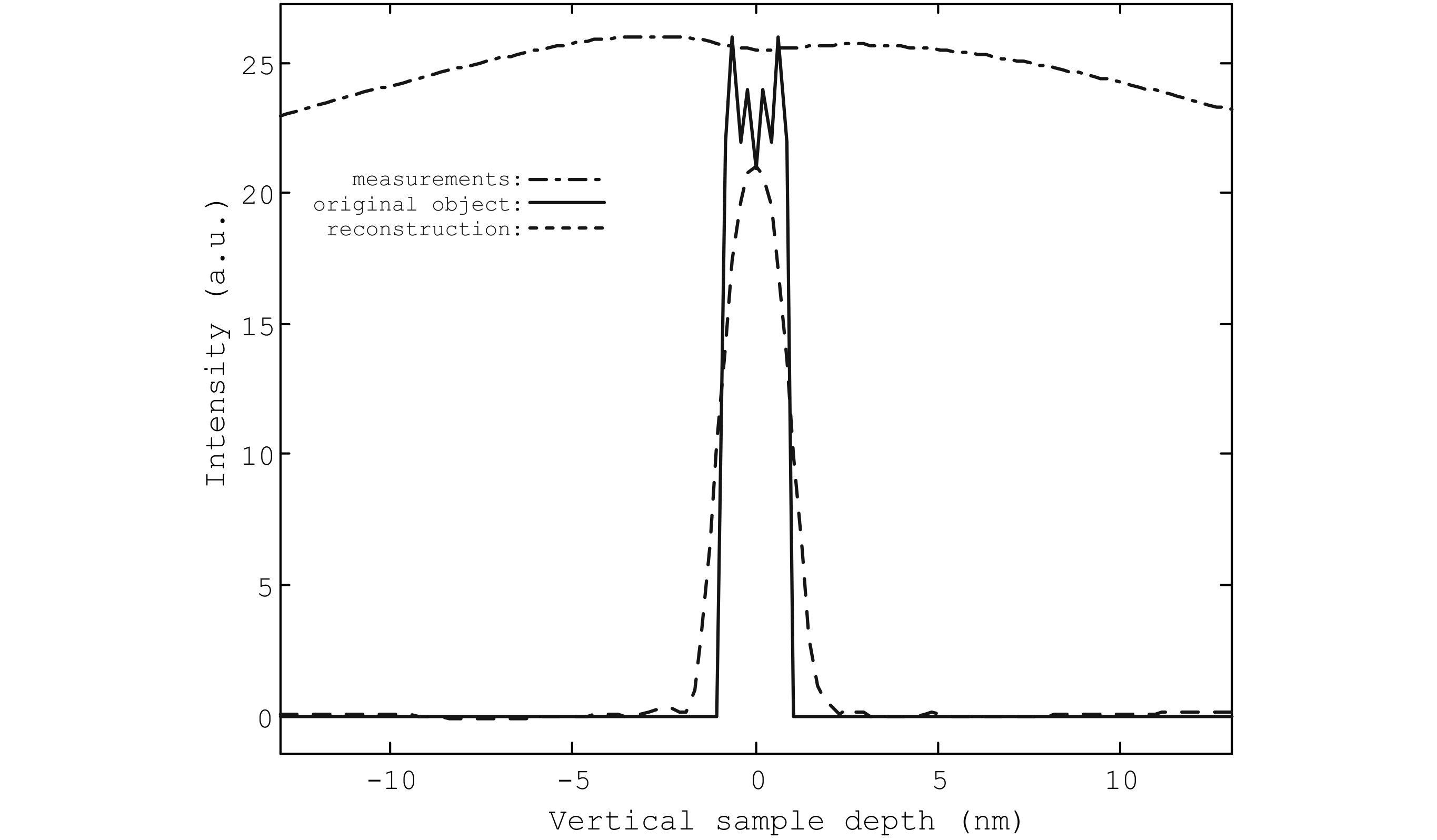
A total of 125 images with a defocus step of 0.21nm is simulated and used as input for SIRT with 64 iterations. In Fig. 3 it is shown that the severe elongation in the vertical direction, so typical for depth sectioning, is overcome. The authors have reported these results in [A memory efficient method for fully three-dimensional object reconstruction with HAADF STEM, Ultram., accepted].
W. Van den Broek: The Carl Zeiss Foundation and DFG, KO 2911/7-1; A. Rosenauer: DFG, AR 2057/8-1; S. Van Aert, J. Sijbers, D. Van Dyck: FWO, G.0393.11, G.0064.10, G.0374.13.