IT-10-O-1863 Towards mass contrast in 3D atomic resolution tomographic reconstructions from HRTEM images with the inverse dynamical electron scattering method
We have recently reported a novel reconstruction algorithm, inverse dynamical electron scattering (IDES), that retrieves the object’s electrostatic potential from a high resolution transmission electron microscopy (HRTEM) tilt series [1]. By reformulating the multislice algorithm as an artificial neural network IDES is able to very efficiently recover the scattering object using a gradient based optimization. Prior knowledge about the atomic potential shape and the sparseness of the object is accounted for naturally. In [2] we demonstrated the generality of this method with applications to simulated ptychography and scanning confocal electron microscopy data sets and the optimization of the images’ defocus values along with the object.
Reconstructions so far had been performed on simulations of a small Au nanoparticle that was 1.6nm wide. In this work a larger and more complicated system is treated: A PbSe-CdSe core-shell particle with 1963 atoms. The particle is derived from a CdSe rock-salt structure and has a cubic shape with sides of 3.7nm. The unit-cell parameter a equals 0.61nm. The Cd atoms in the cube’s interior octahedron are replaced by Pb-atoms and shifted by a vector [−a/4, a/4, a/4]; see [3].
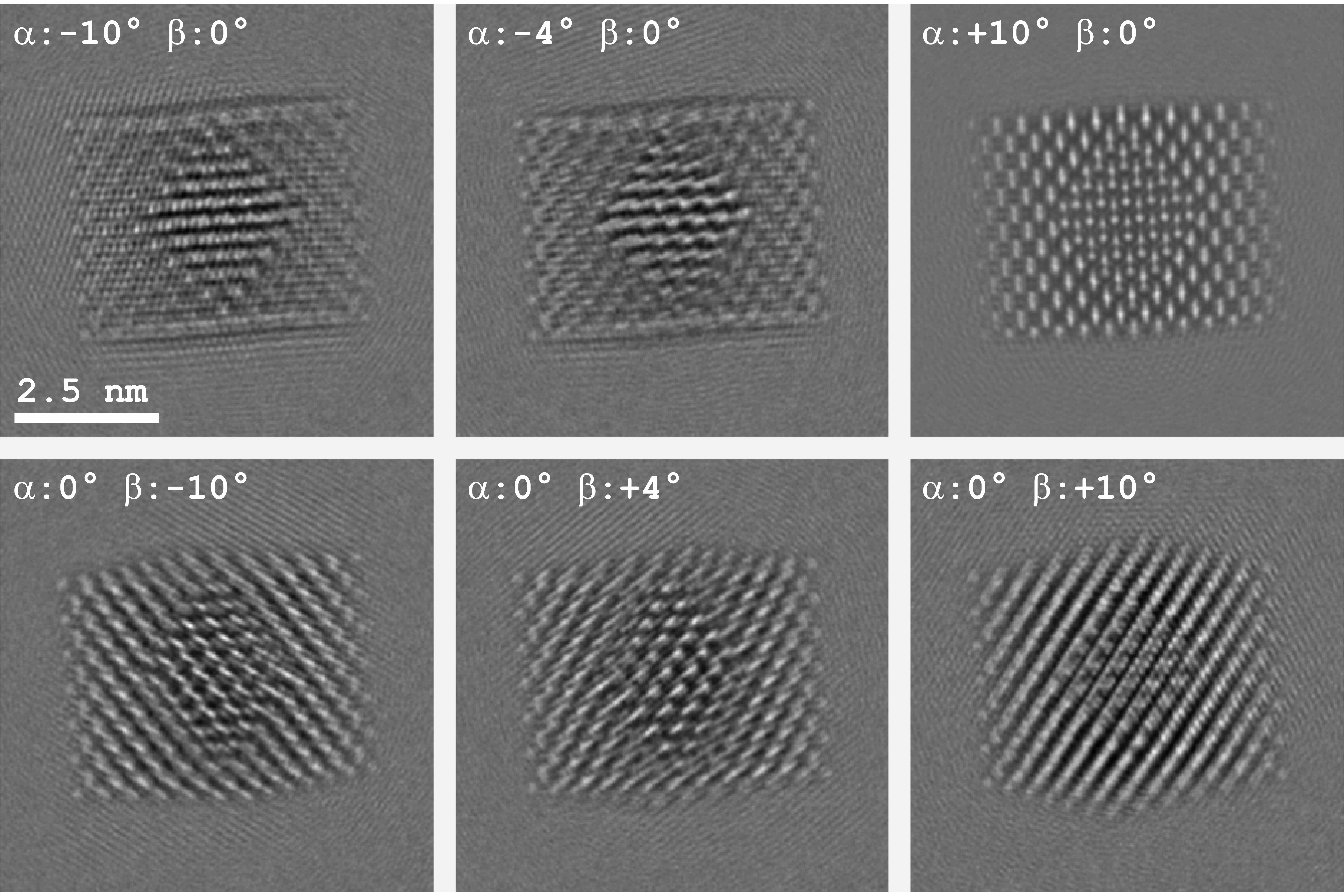
The potential is reconstructed from a simulated HRTEM tilt series with the α-tilt varying from -10° to +10° in 2°-steps with zero β-tilt, and the β-tilt varying from -10° to +10° in 2°-steps with zero α-tilt. The images were given a signal-to-noise ratio of 10. See Fig. 1 for 6 typical simulations out of the total of 21.
The forward simulations have been carried out with the new FDES (forward dynamical electron diffraction) software that runs on the graphics processing unit. A solution was achieved with conjugate gradients optimization. The solution’s sparseness was increased through L1-norm regularization and through the use of a generalized potential, taken as the weighted average of the potentials of Se, Cd and Pb.
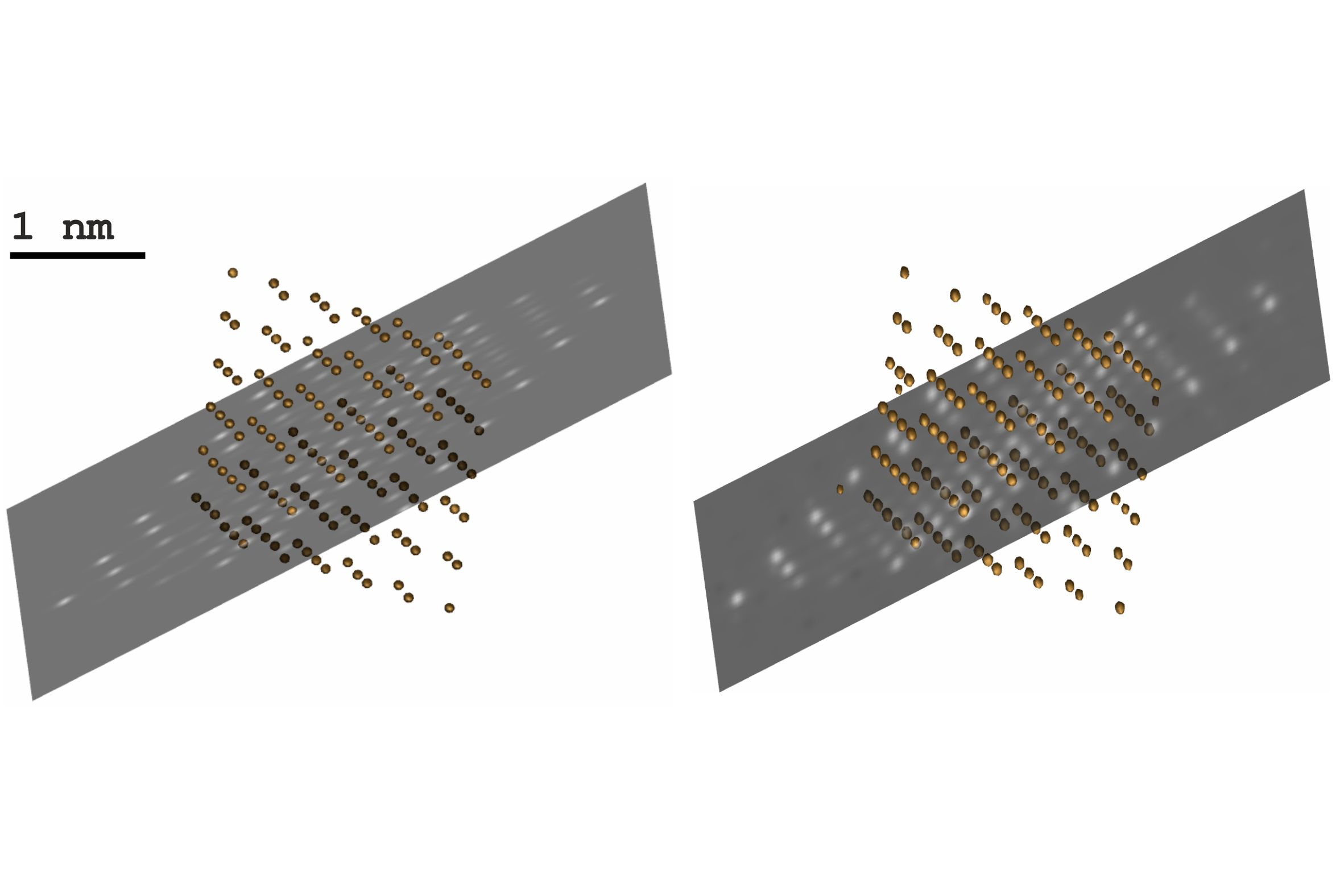
Although FDES was set to use more accurate approximations than IDES—specimen rotation is treated exactly in FDES, but is approximated by a shifted Fresnel propagator in IDES and FDES used a multislice slice thickness 5 times smaller than IDES—an atomic resolution reconstruction was still possible. The Pb-atoms can be isolated by thresholding the reconstructed gray values, thus demonstrating mass contrast. Fig. 2 shows a side-by-side comparison of the original and the reconstructed particle potential, visualized with IMOD [4].
[1] W. Van den Broek, C.T. Koch. Phys. Rev. Lett. 109 (2012) 245502.
[2] W. Van den Broek, C.T. Koch. Phys. Rev. B 87 (2013) 184108.
[3] S. Bals et al. Nano Lett. 11 (2011) 3420–3424.
[4] http://bio3d.colorado.edu/imod/index.html
The authors acknowledge the Carl Zeiss Foundation as well as the German Research Foundation (DFG, Grant No. KO 2911/7-1).